demo_basic
Ivan Selesnick
NYU Tandon School of Engineering
2012, Revised 2016
Contents
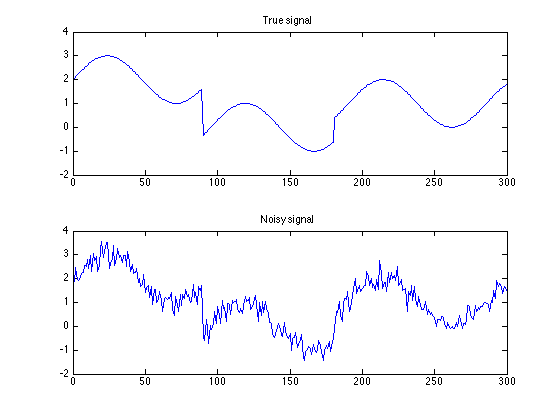
Create test data
clear
N = 300;
n = (1:N)';
s = load('data/basic_signal.txt');
y = load('data/basic_signal_noisy.txt');
Plots
ax = [0 N -2 4];
figure(1)
clf
subplot(2, 1, 1)
plot(s)
axis(ax)
title('True signal');
subplot(2, 1, 2)
plot(y)
axis(ax)
title('Noisy signal');
print -dpdf figures/demo_basic_data
Define low-pass filter
d = 2;
fc = 0.022;
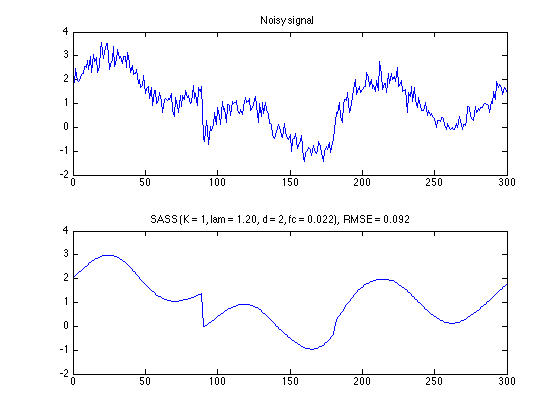
SASS - run algorithm
K = 1;
lam = 1.2;
[x, u] = sass_L1(y, d, fc, K, lam);
err = s - x;
RMSE = sqrt(mean(err.^2));
0 incorrectly locked zeros detected.
SASS - plot denoised signal
txt = sprintf('SASS (K = %d, lam = %.2f, d = %d, fc = %.3f), RMSE = %.3f', K, lam, d, fc, RMSE);
figure(1)
clf
subplot(2, 1, 1)
plot(n, y)
title('Noisy signal');
axis(ax)
subplot(2, 1, 2)
plot(n, x)
title(txt)
axis(ax)
print -dpdf figures/demo_basic_SASS
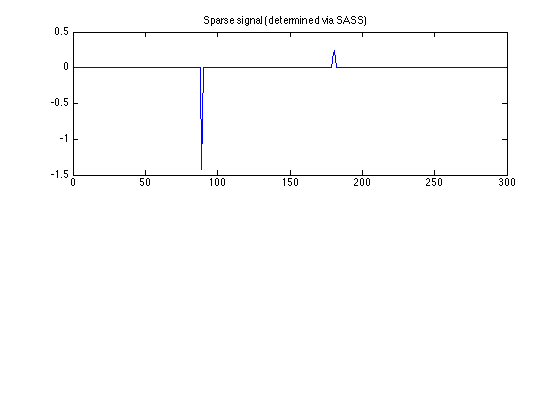
SASS - sparse signal component
figure(1)
clf
subplot(2, 1, 1)
plot(u)
title('Sparse signal (determined via SASS)')
xlim([0 N])
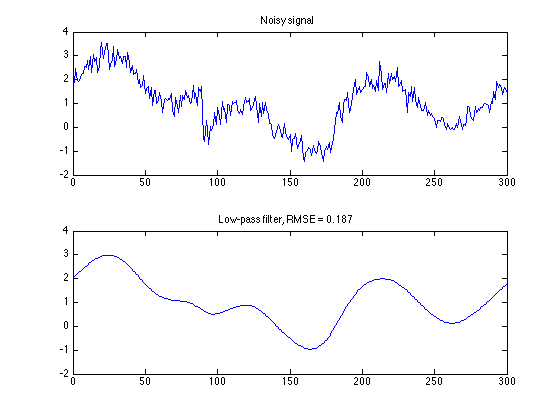
Compare to low-pass filter
[A, B] = ABfilt(d, fc, N, K);
H = @(x) A\(B*x);
L = @(x) x - H(x);
x_lpf = L(y);
err = s - x_lpf;
RMSE_lpf = sqrt(mean(err.^2));
txt_lpf = sprintf('Low-pass filter, RMSE = %.3f', RMSE_lpf);
figure(1)
clf
subplot(2, 1, 1)
plot(n, y)
title('Noisy signal');
axis(ax)
subplot(2, 1, 2)
plot(n, x_lpf)
title(txt_lpf)
axis(ax)
print -dpdf figures/demo_basic_lowpass_filter